My papers
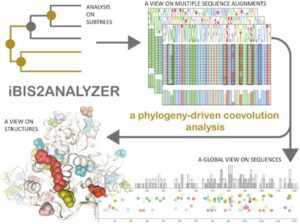
iBIS2Analyzer: a web server for a phylogeny-driven coevolution analysis of protein families.
Oteri, F., Sarti, E., Nadalin, F., & Carbone, A. (2022).
Nucleic Acids Research, 50(W1), W412–W419.
Read the paper
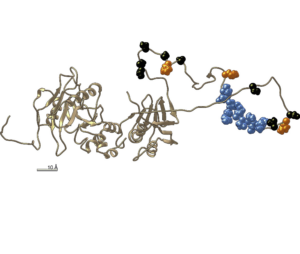
A large disordered region confers a wide spanning volume to vertebrate Suppressor of Fused as shown in a trans-species solution study.
Makamte, S., Thureau, A., Jabrani, A., Paquelin, A., Plessis, A., Sanial, M., Rudenko, O., Oteri, F., Baaden, M., & Biou, V. (2022).
Journal of Structural Biology, 214(2), 107853.
Read the paper
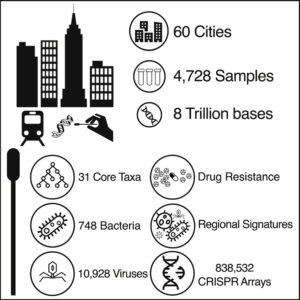
A global metagenomic map of urban microbiomes and antimicrobial resistance.
Danko, D., Bezdan, D., Afshin, E. E., Ahsanuddin, S., Bhattacharya, C., Butler, D. J., Chng, K. R., Donnellan, D., Hecht, J., Jackson, K., Kuchin, K., Karasikov, M., Lyons, A., Mak, L., Meleshko, D., Mustafa, H., Mutai, B., Neches, R. Y., Ng, A., … Zubenko, S. (2021).
Cell, 184(13), 3376-3393.e17.
Read the paper
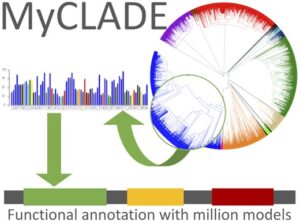
MyCLADE: a multi-source domain annotation server for sequence functional exploration.
Vicedomini, R., Blachon, C., Oteri, F., & Carbone, A. (2021).
Nucleic Acids Research, 49(W1), W452–W458.
Read the paper
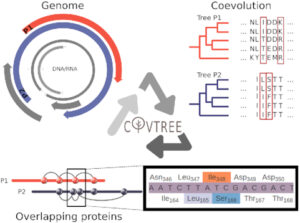
COVTree: Coevolution in OVerlapped sequences by Tree analysis server.
Teppa, E., Zea, D. J., Oteri, F., & Carbone, A. (2020).
Nucleic Acids Research, 48(W1), W558–W565.
Read the paper
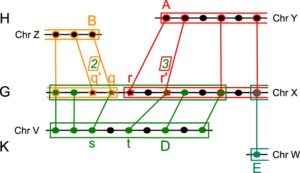
Phylogenetic Reconstruction Based on Synteny Block and Gene Adjacencies.
Drillon, G., Champeimont, R., Oteri, F., Fischer, G., & Carbone, A. (2020).
Molecular Biology and Evolution, 37(9), 2747–2762.
Read the paper
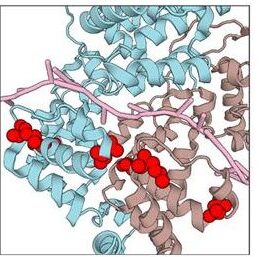
BIS2Analyzer: a server for co-evolution analysis of conserved protein families.
Oteri, F., Nadalin, F., Champeimont, R., & Carbone, A. (2017).
Nucleic Acids Research, 45(W1), W307–W314.
Read the paper
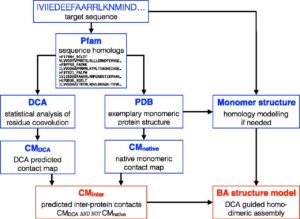
Large-scale identification of coevolution signals across homo-oligomeric protein interfaces by direct coupling analysis.
Uguzzoni, G., John Lovis, S., Oteri, F., Schug, A., Szurmant, H., & Weigt, M. (2017).
Proceedings of the National Academy of Sciences, 114(13).
Read the paper
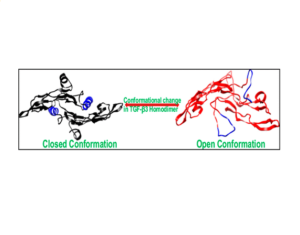
Residues of Alpha Helix H3 Determine Distinctive Features of Transforming Growth Factor β3.
Nayeem, S. M., Oteri, F., Baaden, M., & Deep, S. (2017)
The Journal of Physical Chemistry B, 121(22), 5483–5498.
Read the paper
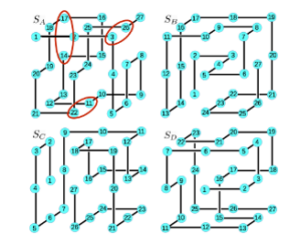
Direct coevolutionary couplings reflect biophysical residue interactions in proteins.
Coucke, A., Uguzzoni, G., Oteri, F., Cocco, S., Monasson, R., & Weigt, M. (2016).
The Journal of Chemical Physics, 145(17), 174102.
Read the paper
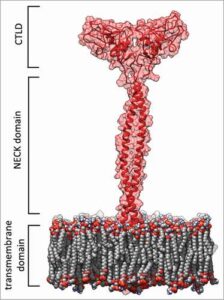
Molecular mechanism of statin-mediated LOX-1 inhibition.
Biocca, S., Iacovelli, F., Matarazzo, S., Vindigni, G., Oteri, F., Desideri, A., & Falconi, M. (2015).
Cell Cycle, 14(10), 1583–1595.
Read the paper
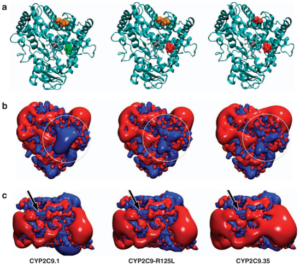
High warfarin sensitivity in carriers of CYP2C9*35 is determined by the impaired interaction with P450 oxidoreductase.
Lee, M.-Y., Borgiani, P., Johansson, I., Oteri, F., Mkrtchian, S., Falconi, M., & Ingelman-Sundberg, M. (2013).
The Pharmacogenomics Journal, 14(4), 343–349.
Read the paper
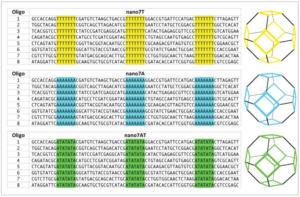
Influence of the single-strand linker composition on the structural/dynamical properties of a truncated octahedral DNA nano-cage family.
Iacovelli, F., Alves, C., Falconi, M., Oteri, F., de Oliveira, C. L. P., & Desideri, A. (2014).
Biopolymers, 101(10), 992–999.
Read the paper
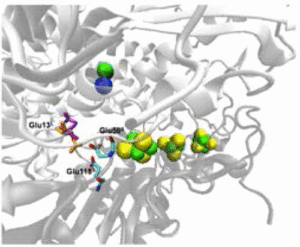
Multiscale Simulations Give Insight into the Hydrogen In and Out Pathways of [NiFe]-Hydrogenases from Aquifex aeolicus and Desulfovibrio fructosovorans.
Oteri, F., Baaden, M., Lojou, E., & Sacquin-Mora, S. (2014).
The Journal of Physical Chemistry B, 118(48), 13800–13811.
Read the paper
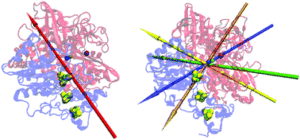
The weak, fluctuating, dipole moment of membrane-bound hydrogenase from Aquifex aeolicus accounts for its adaptability to charged electrodes.
Oteri, F., Ciaccafava, A., Poulpiquet, A. de, Baaden, M., Lojou, E., & Sacquin-Mora, S. (2014).
Phys. Chem. Chem. Phys., 16(23), 11318–11322.
Read the paper
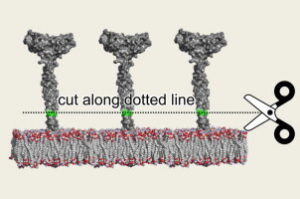
Simulative and experimental investigation on the cleavage site that generates the soluble human LOX-1.
Biocca, S., Arcangeli, T., Tagliaferri, E., Testa, B., Vindigni, G., Oteri, F., Giorgi, A., Iacovelli, F., Novelli, G., Desideri, A., & Falconi, M. (2013).
Archives of Biochemistry and Biophysics, 540(1–2), 9–18.
Read the paper
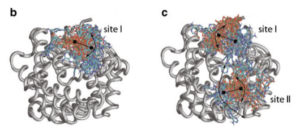
Mapping multiple potential ATP binding sites on the matrix side of the bovine ADP/ATP carrier by the combined use of MD simulation and docking.
Di Marino, D., Oteri, F., Morozzo della Rocca, B., D’Annessa, I., & Falconi, M. (2011).
Journal of Molecular Modeling, 18(6), 2377–2386.
Read the paper
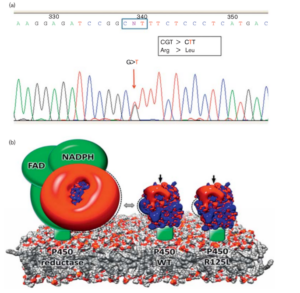
Characterization of a novel CYP2C9 gene mutation and structural bioinformatic protein analysis in a warfarin hypersensitive patient
Ciccacci, C., Falconi, M., Paolillo, N., Oteri, F., Forte, V., Novelli, G., Desideri, A., & Borgiani, P. (2011).
Pharmacogenetics and Genomics, 21(6), 344–346.
Read the paper
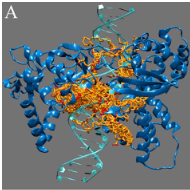
Conjugated Eicosapentaenoic Acid (cEPA) Inhibits L. donovani Topoisomerase I and has an Antiproliferative Activity Against L. donovani Promastigotes.
Vassallo O., Castelli S., Biswas A., Sengupta S., Das P. K., D’Annessa I., Oteri F., Leoni A., Tagliatesta P., Majumder H.K., Desideri A.(2011).
The Open Antimicrobial Agents Journal, 3(1), 23–29.
Read the paper
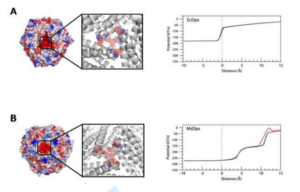
Effect of the charge distribution along the “ferritin-like” pores of the proteins from the Dps family on the iron incorporation process.
Ceci, P., Di Cecca, G., Falconi, M., Oteri, F., Zamparelli, C., & Chiancone, E. (2011).
JBIC Journal of Biological Inorganic Chemistry, 16(6), 869–880.
Read the paper

Simulative Analysis of a Truncated Octahedral DNA Nanocage Family Indicates the Single-Stranded Thymidine Linkers as the Major Player for the Conformational Variability.
Oteri, F., Falconi, M., Chillemi, G., Andersen, F. F., Oliveira, C. L. P., Pedersen, J. S., Knudsen, B. R., & Desideri, A. (2011).
The Journal of Physical Chemistry C, 115(34), 16819–16827.
Read the paper
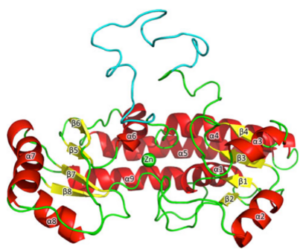
Structural-dynamical investigation of the ZnuA histidine-rich loop: involvement in zinc management and transport.
Falconi, M., Oteri, F., Di Palma, F., Pandey, S., Battistoni, A., & Desideri, A. (2011).
Journal of Computer-Aided Molecular Design, 25(2), 181–194.
Read the paper
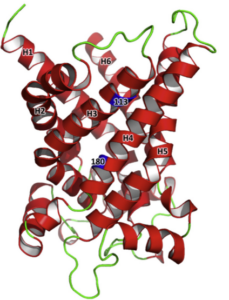
ADP/ATP mitochondrial carrier MD simulations to shed light on the structural–dynamical events that, after an additional mutation, restore the function in a pathological single mutant.
Di Marino, D., Oteri, F., Morozzo della Rocca, B., Chillemi, G., & Falconi, M. (2010).
Journal of Structural Biology, 172(3), 225–232.
Read the paper
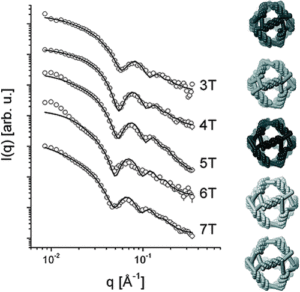
Structure of Nanoscale Truncated Octahedral DNA Cages: Variation of Single-Stranded Linker Regions and Influence on Assembly Yields.
Oliveira, C. L. P., Juul, S., Jørgensen, H. L., Knudsen, B., Tordrup, D., Oteri, F., Falconi, M., Koch, J., Desideri, A., Pedersen, J. S., Andersen, F. F., & Knudsen, B. R. (2010).
ACS Nano, 4(3), 1367–1376.
Read the paper
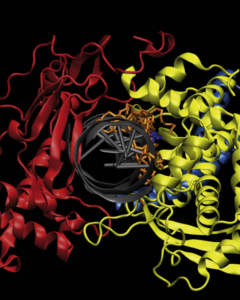
Conjugated eicosapentaenoic acid inhibits human topoisomerase IB with a mechanism different from camptothecin.
Castelli, S., Campagna, A., Vassallo, O., Tesauro, C., Fiorani, P., Tagliatesta, P., Oteri, F., Falconi, M., Majumder, H. K., & Desideri, A. (2009).
Archives of Biochemistry and Biophysics, 486(2), 103–110.
Read the paper
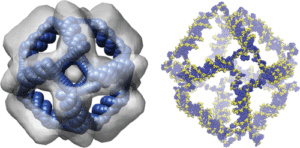
Deciphering the Structural Properties That Confer Stability to a DNA Nanocage.
Falconi, M., Oteri, F., Chillemi, G., Andersen, F. F., Tordrup, D., Oliveira, C. L. P., Pedersen, J. S., Knudsen, B. R., & Desideri, A. (2009).
ACS Nano, 3(7), 1813–1822.
Read the paper
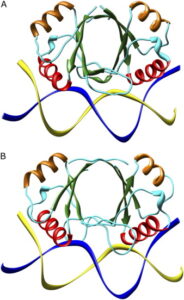
MD Simulations of Papillomavirus DNA-E2 Protein Complexes Hints at a Protein Structural Code for DNA Deformation.
Falconi, M., Oteri, F., Eliseo, T., Cicero, D. O., & Desideri, A. (2008).
Biophysical Journal, 95(3), 1108–1117.
Read the paper